
Looking into the diagnostic workup of dedifferentiated liposarcoma (DDLPS)
Due to their low prevalence and heterogeneity, sarcomas present challenges for accurate diagnosis1,2
The histologic hallmark of DDLPS is the abrupt transition from WDLPS to DDLPS3
- DDLPS can be associated with well-defined areas; therefore, in a diagnosis, distinguishing DDLPS from WDLPS is important2Biomarkers are emerging as an important component of diagnosis for DDLPS:
- MDM2: Distinguishes DDLPS and WDLPS from benign lipomas4
- CDK4: Co-amplification with MDM2 is a sensitive and specific marker for diagnosis of DDLPS/WDLPS4
- HMGA2: Amplified in 60% of DDLPS cases4
- CPM, YEATS4: Implicated in dedifferentiation5
Getting deeper into the diagnostic workup of DDLPS
| Tumour biopsy | Biomarker testing |
|---|---|
Tumour biopsy helps to determine histologic type and subtype, as well as the extent of tumour necrosis6
To complement the histological analysis, cross-sectional imaging based on tumour location assists in the establishment of differential diagnosis6 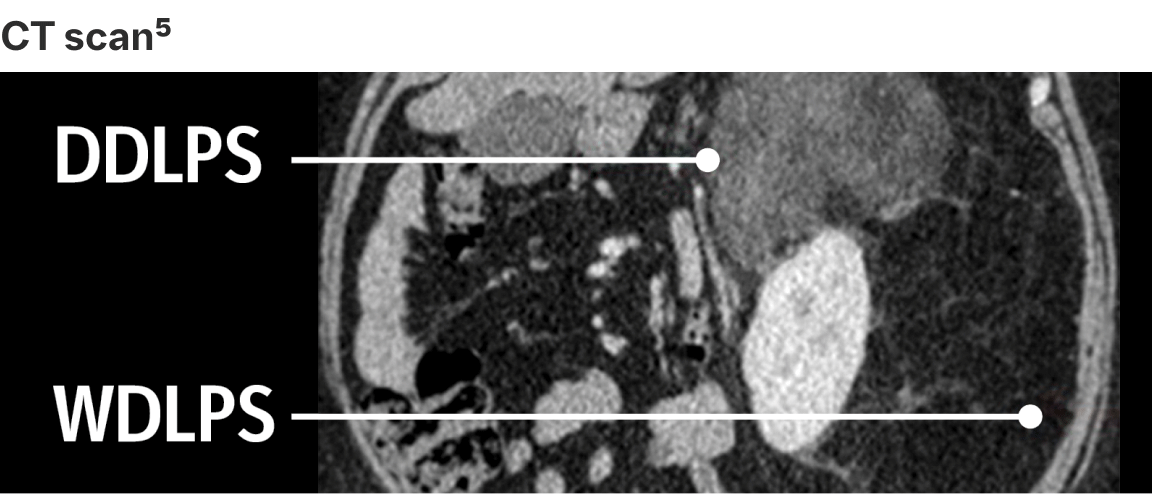
| Excess MDM2 is a hallmark of DDLPS; nearly all patients with advanced DDLPS present with excess MDM28
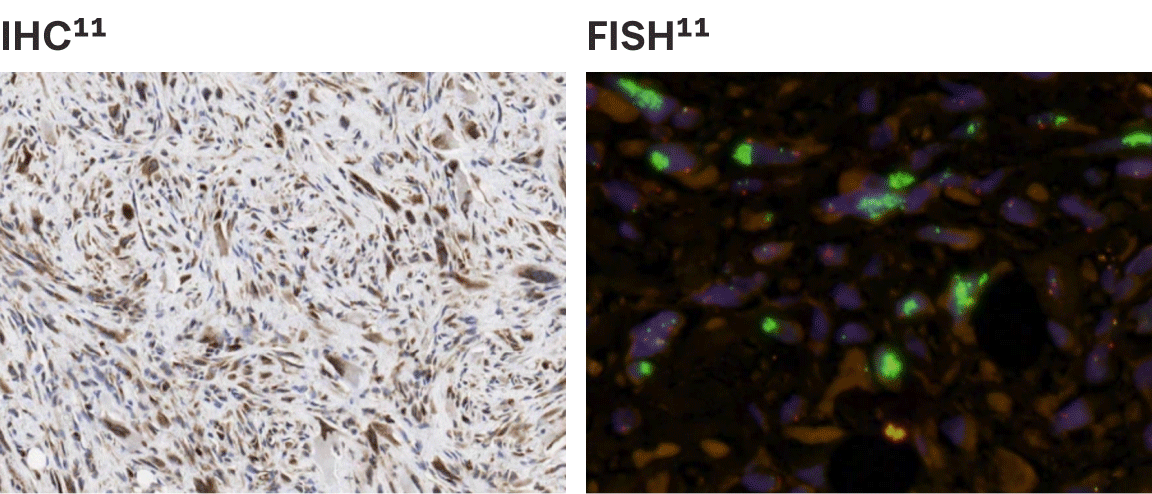
|
DDLPS belongs to a heterogeneous group of rare tumours where management by a group of experienced specialists is suggested in patient care from the time of diagnosis6

Retroperitoneal tumours present later; due to their anatomical location, they often remain physically hidden, resulting in delayed diagnosis2,12
Given the complexities of DDLPS, what is the best approach to managing this sarcoma?
Go to the next page to learn the recommended approach.
CDK4=cyclin-dependent kinase 4; CPM=carboxypeptidase M; FISH=fluorescence in situ hybridisation; HMGA2=high-mobility group AT-hook 2; IHC=immunohistochemistry; MDM2=mouse double minute 2; NGS=next-generation sequencing; WDLPS=well-differentiated liposarcoma; YEATS4=YEATS domain-containing protein 4.
References:
-
Gounder MM, Agaram NP, Trabucco SE, et al. Nat Commun. 2022;13(1):3406. doi:10.1038/s41467-022-30496-0
-
Gahvari Z, Parkes A. Curr Treat Options Oncol. 2020;21(2):15. doi:10.1007/s11864-020-0705-7
-
Nishio J, Nakayama S, Nabeshima K, Yamamoto T. J Clin Med. 2021;10(15):3230. doi:10.3390/jcm10153230
-
Jagosky MH, Anderson CJ, Symanowski JT, et al. Cancer Med. 2023;12(6):7029-7038.
-
Lee ATJ, Thway K, Huang PH, Jones RL. J Clin Oncol. 2018;36(2):151-159. doi:10.1200/JCO.2017.74.9598
-
Gamboa AC, Gronchi A, Cardona K. CA Cancer J Clin. 2020;70(3):200-229.
-
Coindre JM, Pédeutour F, Aurias A. Virchows Arch. 2010;456(2):167-179.
-
McGovern Y, Zhou CD, Jones RL. Front Oncol. 2017;7:292. doi:10.3389/fonc.2017.00292
-
Thway K. Semin Diagn Pathol. 2019;36(2):112-121.
-
Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Soft Tissue Sarcoma V.3.2023. © National Comprehensive Cancer Network, Inc. 2023. All rights reserved. Accessed 6 March 2024. To view the most recent and complete version of the guidelines, go online to NCCN.org. NCCN makes no warranties of any kind whatsoever regarding their content, use or application and disclaims any responsibility for their application or use in any way.
-
Lokka S, Scheel AH, Dango S, et al. BMC Clin Pathol. 2014;14:36. doi:10.1186/1472-6890-14-3
-
Nguyen K, Gootee J, Aurit S, Albagoush S, Curtin C, Silberstein P. Surg Case Rep. 2021;4(2):7-7. doi:10.31487/j.SCR.2021.02.02